It is day four since our departure, and I have been busy filtering water samples from two of the sites we extensively surveyed and sampled; “Virginia Inter-Canyon” and “Hatteras South”. While there are other findings (e.g. size and presence of organic matter particulates) you can obtain in filtering material from the water column, my goal is to test whether we can determine the taxonomic composition of the organisms in the water column based on the environmental DNA (eDNA) collected from the water on a filter.
An intuitive definition of eDNA is simply DNA that can be extracted from bulk environmental samples such as soil, sediments in aquatic systems, and in this case water. This eDNA can consist of sloughed cells, excretions, parts of organisms, or in the case of really small things like diatoms and bacteria, the whole organism. When filtering water for eDNA, the pore size of the filter used determines what material is captured for a DNA extraction and analysis. Pictured below is a filter capsule housing a 0.2 micron filter. A peristaltic pump is used to suction the water sample from a container and push it through this filter, which retains material larger than the 0.2 micron pore size. A 0.2 micron filter is generally assumed to retain things the size of most bacteria and larger. I am filtering 1 liter samples of seawater collected from throughout the water column through these filters, and then keeping these filters frozen to protect the retained particulate from degradation until we return to land and can analyze the samples in the lab.
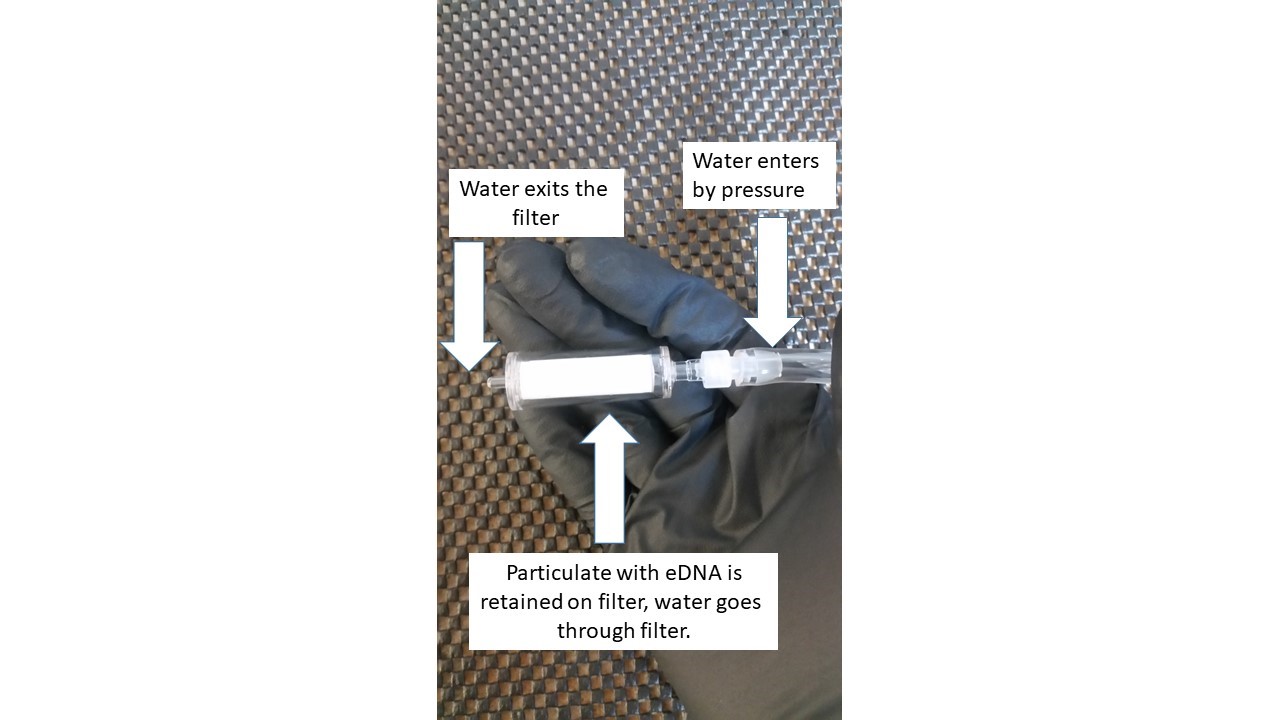
From this “soup” of DNA (diatoms, fishes, bacteria, etc…) on the filter, one can target specific taxa for sequencing using primers that pick up all members of that group (hopefully! – stay tuned for another future post about DNA sequencing primer design and building the reference database for a community sequencing study). In the lab, we will be using the sequencing technique of meta-barcoding to hone in on specific taxonomic groups such as fishes and cold water corals. The ultimate prize is to develop a means of recovering the taxa present at a site through simple filtering of a water sample at a level of sensitivity comparable to, or exceeding traditional methods such as trawling or visualizing the bottom with an ROV. This new method requires just a few water samples and would be useful for surveying broad geographic areas in situations where other sampling methods are not feasible, or as a quick means of surveying difficult to reach deep habitats.
By Aaron Aunins, USGS.


